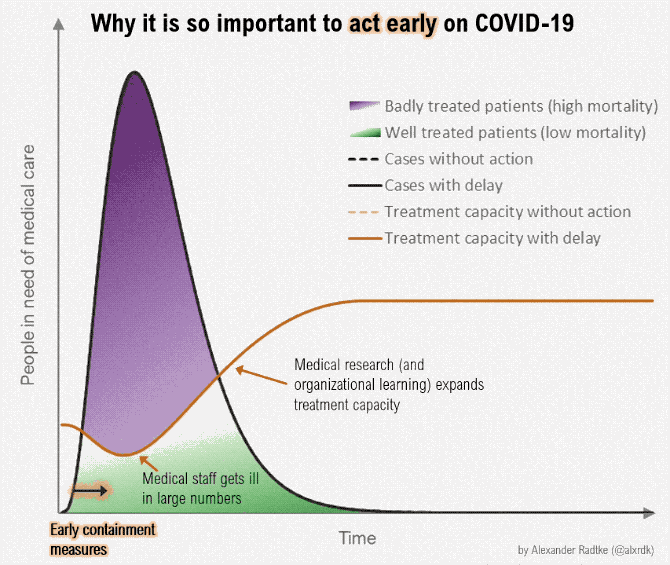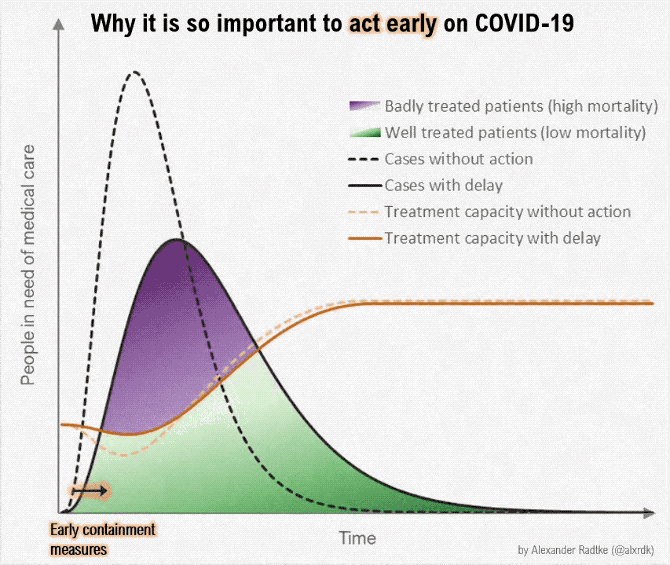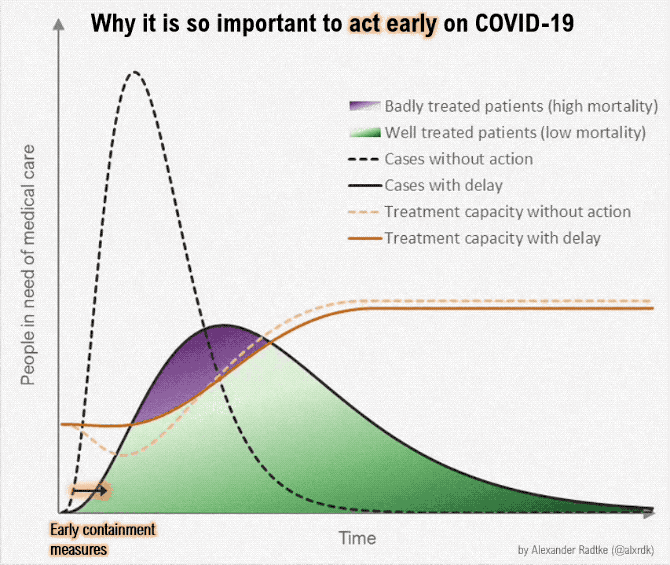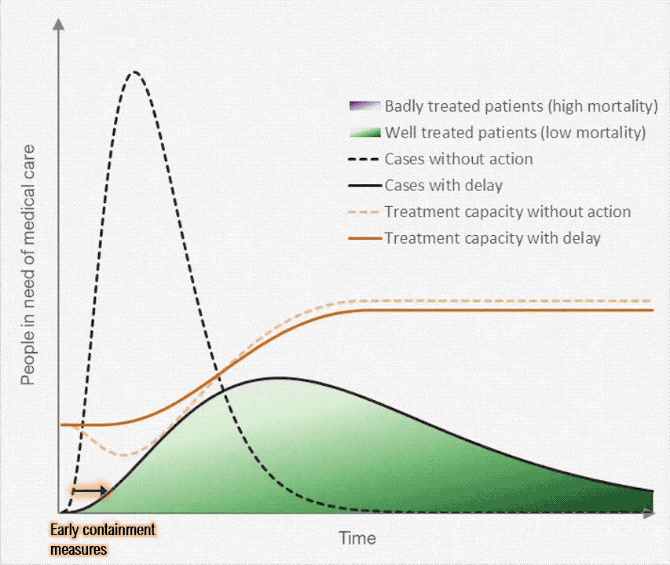
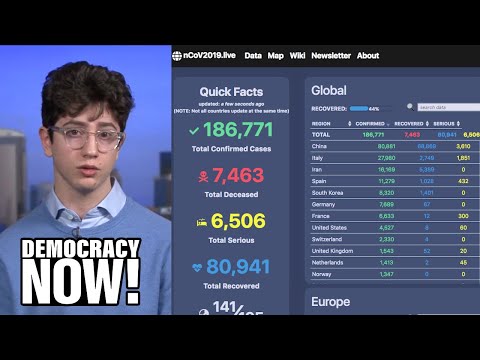
Gezien alle fake news en onzin die wordt geplaatst over het Coronavirus, plaats hier alle relevante peer reviewed wetenschappelijke artikelen over Corona (of ook andere artikelen/interviews zolang het door een dokter/wetenschapper is gemaakt). Gelukkig is er veel onderzoek gedaan naar het Coronavirus.
Plaats ook verschillende data analyses/visualisaties en statistieken over het Corona virus.
Actuele informatie en data van het RIVM, dagelijkse update om 14:00
WHO Corona website
Elsevier corona information centre
Coronavirus laatste nieuws van BNO
Aantal bekende grafieken:
Plaats ook verschillende data analyses/visualisaties en statistieken over het Corona virus.
Algemene linkjesquote:SARS-CoV-2, voorheen bekend als 2019-nCoV, is een virus dat de besmettelijke luchtwegaandoening COVID-19 bij mensen veroorzaakt.SARS-CoV-2 is een virus-stam uit het geslacht betacoronavirussen uit de onderfamilie van coronavirussen.
Actuele informatie en data van het RIVM, dagelijkse update om 14:00
WHO Corona website
Elsevier corona information centre
Coronavirus laatste nieuws van BNO
Aantal bekende grafieken:
SPOILEROm spoilers te kunnen lezen moet je zijn ingelogd. Je moet je daarvoor eerst gratis Registreren. Ook kun je spoilers niet lezen als je een ban hebt.Data visualisaties:
https://twitter.com/Datagraver
Voorbeelden:SPOILEROm spoilers te kunnen lezen moet je zijn ingelogd. Je moet je daarvoor eerst gratis Registreren. Ook kun je spoilers niet lezen als je een ban hebt.Worldometer coronavirus site
Mapping the Outbreak timelapse
Preppers kaart verspreiding in NL Nog een kaartje van NL
Nog een kaartje van NL
Wuhan Corona Virus Map
Livetrackers:
(link 1) (link 2) (link 3)
Wetenschappelijke artikelen:
A pneumonia outbreak associated with a new coronavirus of probable bat origin
Coronavirus latest: First vaccine clinical trials begin in United States
China’s response to a novel coronavirus stands in stark contrast to the 2002 SARS outbreak response
Arguments in favor of remdesivir for treating SARS-CoV-2 infections
Chloroquine and hydroxychloroquine as available weapons to fight COVID-19
Coronavirus Disease 2019 (COVID-19): Protecting Hospitals From the Invisible
Clinical features of deaths in the novel coronavirus epidemic in China
COVID‐19 and Rationally Layered Social Distancing
Mental health groups providing support, education in wake of COVID‐19
The potential chemical structure of anti‐SARS‐CoV‐2 RNA‐dependent RNA polymerase
A report of clinical diagnosis and treatment of nine cases of coronavirus disease 2019
2019 novel coronavirus patients’ clinical characteristics, discharge rate and fatality rate of meta‐analysis
Novel Coronavirus (COVID‐19) Epidemic: What Are the Risks for Older Patients?
Plaats gerust artikelen en data en andere verhalen over de mortality rates, comorbidity, comortality, besmettelijkheid, medicijnen, vaccinaties, behandelingen,aanpak van overheden, mentale gezondheid etc. En data visualisaties/analyse en andere statistieken over het Coronavirus. Bij interessante bronnen en nieuws probeer ik de OP te updaten. Deel vooral hier, en zet er altijd even een bron bij. Geen fake news of sensatie nieuws van kranten hier aub
[ Bericht 0% gewijzigd door Momo op 17-03-2020 17:28:48 ]
Also known as Kyran or Theoden


Goed idee. Alleen denk ik dat er te weinig peer reviewed artikelen zullen zijn de komende tijd. Misschien uitbreiden naar ook niet peer reviewed artikelen?
Gewoon een idee
Gewoon een idee


Ja zolang het wetenschappelijke inhoud heeft, kan ook een interview gewoon zijn met een expert.quote:Op dinsdag 17 maart 2020 17:03 schreef Kyran het volgende:
Goed idee. Alleen denk ik dat er te weinig peer reviewed artikelen zullen zijn de komende tijd. Misschien uitbreiden naar ook niet peer reviewed artikelen?
Gewoon een idee


quote:Fosun Pharma and BioNTech form COVID-19 vaccine strategic alliance in China
(15 March 2020, Shanghai, Hong Kong) – Today, Fosun Pharma industrial, a subsidiary company of Shanghai Fosun Pharmaceutical (Group) Co., Ltd (“Fosun Pharma” or “the Group”; Stock Code: 600196.SH, 02196.HK) and BioNTech SE (NASDAQ: BNTX, “BioNTech” or “the Company”) announced a strategic development and commercialization collaboration to advance BioNTech’s mRNA vaccine in China for the prevention of COVID-19 infections.
Under the terms of the agreement, the two Companies will work jointly on the development of COVID-19 vaccines based on BioNTech’s mRNA technology platform in China. The Companies will collaborate to conduct clinical trials in China leveraging Fosun Pharma’s extensive clinical development, regulatory, and commercial capabilities in the country. BioNTech will supply the mRNA vaccine for clinical trials from GMP manufacturing facilities in Europe.
mRNA is a nucleic acid molecule that carries genetic information. The mRNA vaccine introduces the genetic information into the body, so that the cells in the body produce the corresponding antigen, which induces the body to produce neutralizing antibodies and stimulates the response of T cells, and fights against the virus through the dual mechanisms of humoral immunity and cellular immunity. In the face of a sudden epidemic, compared with traditional vaccines, synthesis and production process of mRNA vaccine is more convenient, and has strong immunogenicity, which does not need additional adjuvant as needed by the traditional vaccine, and with good safety.
BioNTech is a leading mRNA technology company that has diversified mRNA platform technologies, integrated R&D system and strong manufacturing capability. “The mRNA technology introduces genetic information into human body, then the body's own cells produce the corresponding proteins that cure or prevent disease. Fosun Pharma R&D has been tracking the development of this technology. The two companies expect further cooperation in this field,” says Dr. Aimin Hui, President of Fosun Pharma Global R&D.
“We see this collaboration as an important step in our global effort to expedite the development of mRNA vaccines to prevent COVID-19 infection. Fosun Pharma shares our commitment to move rapidly to address the COVID-19 outbreak and brings deep development experience and an extensive network in the pharmaceutical market in China,” says Founder and CEO of BioNTech, Ugur Sahin, M.D.
Wu Yifang, President and CEO of Fosun Pharma states, “A potential pandemic requires a collective effort and both companies are passionate about contributing to the fight against the current coronavirus outbreak. We are excited to collaborate with BioNTech, one of the leading companies worldwide in the mRNA field. Our shared objective is to develop a vaccine against the coronavirus and to be able to rapidly manufacture a vaccine to turn the tide of COVID-19 infection.”
Under the terms of the agreement, Fosun Pharma will pay BioNTech up to $85 million in licensing fees (including a down payment, clinical development registration and sales milestone payments) as agreed, and a sales commission equal to 35% of the product's annual gross profit during the agreed sales commission period. Meanwhile, Fosun Pharma has agreed to make an equity investment of USD 50 million (EUR 44 million) for 1,580,777 ordinary shares in BioNTech.


Design of an Epitope-Based Synthetic Long Peptide Vaccine to Counteract the Novel China Coronavirus (2019-nCoV)
In this report, we demonstrate that it is possible to design epitope-based peptide vaccine candidates to counteract the novel China coronavirus (2019-nCoV) by using an approach similar to the one used in cancer neoantigen vaccination therapy. We identified multiepitope peptide vaccine candidates against 2019-nCov that can potentially trigger both CD4+ and CD8+ T cell immune response with increased efficiency due to the presence of CD4+ and CD8+ T cell epitopes and a cathepsin-sensitive linker. Furthermore, we suggest that the peptide design strategy should incorporate population-specific HLA alleles in order to optimize binding specificity of the peptides. We refer to this as populationalized vaccinomics.
In this report, we demonstrate that it is possible to design epitope-based peptide vaccine candidates to counteract the novel China coronavirus (2019-nCoV) by using an approach similar to the one used in cancer neoantigen vaccination therapy. We identified multiepitope peptide vaccine candidates against 2019-nCov that can potentially trigger both CD4+ and CD8+ T cell immune response with increased efficiency due to the presence of CD4+ and CD8+ T cell epitopes and a cathepsin-sensitive linker. Furthermore, we suggest that the peptide design strategy should incorporate population-specific HLA alleles in order to optimize binding specificity of the peptides. We refer to this as populationalized vaccinomics.


Ook even hier een linkje naar het topic van @_I over de verschillende mutaties van Corona virus die nu in de wereld rond gaan. Zo kun je bijvoorbeeld zien dat het Corona virus dat nu vooral actief in de VS vooral uit AziŽ komt en niet uit Europa. De varianten die je vooral in Nederland vind zijn afkomstig uit Noord-ItaliŽ.
Nextstrain
En de website waar dat op staat:
https://nextstrain.org/ncov?c=country&f_country=Japan,USA,China&l=radial
Nextstrain
En de website waar dat op staat:
https://nextstrain.org/ncov?c=country&f_country=Japan,USA,China&l=radial


Zie voorlopig behandeladvies RIVM, en voor behandelopties RIVMquote:Op dinsdag 17 maart 2020 19:42 schreef merdobach het volgende:
Is al bekend welke onderzoeken het rivm gebruikt?
[ Bericht 0% gewijzigd door Momo op 17-03-2020 19:53:30 ]


https://www.overhetcoronavirus.nl/
arts, klinisch epidemioloog en onderzoeker uit Utrecht die hoofdzakelijk wetenschappelijke bronnen gebruikt.
arts, klinisch epidemioloog en onderzoeker uit Utrecht die hoofdzakelijk wetenschappelijke bronnen gebruikt.
Realiteit is een illusie die ontstaat bij een gebrek aan alcohol


How will country-based mitigation measures influence the course of the COVID-19 epidemic?
Hier ander onderzoek van RIVM onderzoekers in samenwerking met 2 Britse onderzoekers die daar ook in een vergelijkbare rol werken. Ook een beetje in lijn met wat Rutte zei over het verspreiden van besmettingen.
Hier ander onderzoek van RIVM onderzoekers in samenwerking met 2 Britse onderzoekers die daar ook in een vergelijkbare rol werken. Ook een beetje in lijn met wat Rutte zei over het verspreiden van besmettingen.


Ik heb geen verstand van het verloop van mutaties bij virussen. Maar is er ook al iets te zeggen of de verschillende aftakkingen zich anders (bijvoorbeeld agressiever of milder) gedragen? Of is het daar nog te vroeg voor? Of zijn de mutaties die in zo'n korte periode optreden Łberhaupt niet significant genoeg?quote:Op dinsdag 17 maart 2020 19:34 schreef Momo het volgende:
Ook even hier een linkje naar het topic van @:_I over de verschillende mutaties van Corona virus die nu in de wereld rond gaan. Zo kun je bijvoorbeeld zien dat het Corona virus dat nu vooral actief in de VS vooral uit AziŽ komt en niet uit Europa. De varianten die je vooral in Nederland vind zijn afkomstig uit Noord-ItaliŽ.
Nextstrain
En de website waar dat op staat:
https://nextstrain.org/ncov?c=country&f_country=Japan,USA,China&l=radial
Als je naar de sterftecijfers in ItaliŽ kijkt dan zou je bijna denken dat daar een meer agressieve variant heerst. Maar dat kan ook hele andere oorzaken hebben (gemiddelde leeftijd van de geÔnfecteerden, worden alle besmettingen vastgesteld/geregistreerd, enzovoort).


https://www.globaltimes.cn/content/1181612.shtmlquote:Op dinsdag 17 maart 2020 20:33 schreef Breekfast het volgende:
[..]
Ik heb geen verstand van het verloop van mutaties bij virussen. Maar is er ook al iets te zeggen of de verschillende aftakkingen zich anders (bijvoorbeeld agressiever of milder) gedragen? Of is het daar nog te vroeg voor? Of zijn de mutaties die in zo'n korte periode optreden Łberhaupt niet significant genoeg?
Als je naar de sterftecijfers in ItaliŽ kijkt dan zou je bijna denken dat daar een meer agressieve variant heerst. Maar dat kan ook hele andere oorzaken hebben (gemiddelde leeftijd van de geÔnfecteerden, worden alle besmettingen vastgesteld/geregistreerd, enzovoort).
Dat vermoeden is er wel. Waarschijnlijk wordt daar nu meer onderzoek naar gedaan.


Theories of SARS-CoV-2 origins
SPOILEROm spoilers te kunnen lezen moet je zijn ingelogd. Je moet je daarvoor eerst gratis Registreren. Ook kun je spoilers niet lezen als je een ban hebt.Nature


The view from nowhere.


Wat betreft de invloed van temperatuur en vochtigheid op de verspreiding van het virus:
High Temperature and High Humidity Reduce the Transmission of COVID-19
This paper investigates how air temperature and humidity influence the transmission of COVID-19. After estimating the serial interval of COVID-19 from 105 pairs of the virus carrier and the infected, we calculate the daily effective reproductive number, R, for each of all 100 Chinese cities with more than 40 cases. Using the daily R values from January 21 to 23, 2020 as proxies of non-intervened transmission intensity, we find, under a linear regression framework for 100 Chinese cities, high temperature and high relative humidity significantly reduce the transmission of COVID-19, respectively, even after controlling for population density and GDP per capita of cities. One degree Celsius increase in temperature and one percent increase in relative humidity lower R by 0.0383 and 0.0224, respectively. This result is consistent with the fact that the high temperature and high humidity significantly reduce the transmission of influenza. It indicates that the arrival of summer and rainy season in the northern hemisphere can effectively reduce the transmission of the COVID-19.
[ Bericht 5% gewijzigd door Momo op 18-03-2020 10:08:53 ]
High Temperature and High Humidity Reduce the Transmission of COVID-19
This paper investigates how air temperature and humidity influence the transmission of COVID-19. After estimating the serial interval of COVID-19 from 105 pairs of the virus carrier and the infected, we calculate the daily effective reproductive number, R, for each of all 100 Chinese cities with more than 40 cases. Using the daily R values from January 21 to 23, 2020 as proxies of non-intervened transmission intensity, we find, under a linear regression framework for 100 Chinese cities, high temperature and high relative humidity significantly reduce the transmission of COVID-19, respectively, even after controlling for population density and GDP per capita of cities. One degree Celsius increase in temperature and one percent increase in relative humidity lower R by 0.0383 and 0.0224, respectively. This result is consistent with the fact that the high temperature and high humidity significantly reduce the transmission of influenza. It indicates that the arrival of summer and rainy season in the northern hemisphere can effectively reduce the transmission of the COVID-19.
[ Bericht 5% gewijzigd door Momo op 18-03-2020 10:08:53 ]


Aerosol and Surface Stability of SARS-CoV-2 as Compared with SARS-CoV-1
A novel human coronavirus that is now named severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) (formerly called HCoV-19) emerged in Wuhan, China, in late 2019 and is now causing a pandemic.1 We analyzed the aerosol and surface stability of SARS-CoV-2 and compared it with SARS-CoV-1, the most closely related human coronavirus.2
We evaluated the stability of SARS-CoV-2 and SARS-CoV-1 in aerosols and on various surfaces and estimated their decay rates using a Bayesian regression model (see the Methods section in the Supplementary Appendix, available with the full text of this letter at NEJM.org). SARS-CoV-2 nCoV-WA1-2020 (MN985325.1) and SARS-CoV-1 Tor2 (AY274119.3) were the strains used. Aerosols (<5 μm) containing SARS-CoV-2 (105.25 50% tissue-culture infectious dose [TCID50] per milliliter) or SARS-CoV-1 (106.75-7.00 TCID50 per milliliter) were generated with the use of a three-jet Collison nebulizer and fed into a Goldberg drum to create an aerosolized environment. The inoculum resulted in cycle-threshold values between 20 and 22, similar to those observed in samples obtained from the upper and lower respiratory tract in humans.
Our data consisted of 10 experimental conditions involving two viruses (SARS-CoV-2 and SARS-CoV-1) in five environmental conditions (aerosols, plastic, stainless steel, copper, and cardboard). All experimental measurements are reported as means across three replicates.
SARS-CoV-2 remained viable in aerosols throughout the duration of our experiment (3 hours), with a reduction in infectious titer from 103.5 to 102.7 TCID50 per liter of air. This reduction was similar to that observed with SARS-CoV-1, from 104.3 to 103.5 TCID50 per milliliter (Figure 1A).
SARS-CoV-2 was more stable on plastic and stainless steel than on copper and cardboard, and viable virus was detected up to 72 hours after application to these surfaces (Figure 1A), although the virus titer was greatly reduced (from 103.7 to 100.6 TCID50 per milliliter of medium after 72 hours on plastic and from 103.7 to 100.6 TCID50 per milliliter after 48 hours on stainless steel). The stability kinetics of SARS-CoV-1 were similar (from 103.4 to 100.7 TCID50 per milliliter after 72 hours on plastic and from 103.6 to 100.6 TCID50 per milliliter after 48 hours on stainless steel). On copper, no viable SARS-CoV-2 was measured after 4 hours and no viable SARS-CoV-1 was measured after 8 hours. On cardboard, no viable SARS-CoV-2 was measured after 24 hours and no viable SARS-CoV-1 was measured after 8 hours (Figure 1A).
Both viruses had an exponential decay in virus titer across all experimental conditions, as indicated by a linear decrease in the log10TCID50 per liter of air or milliliter of medium over time (Figure 1B). The half-lives of SARS-CoV-2 and SARS-CoV-1 were similar in aerosols, with median estimates of approximately 1.1 to 1.2 hours and 95% credible intervals of 0.64 to 2.64 for SARS-CoV-2 and 0.78 to 2.43 for SARS-CoV-1 (Figure 1C, and Table S1 in the Supplementary Appendix). The half-lives of the two viruses were also similar on copper. On cardboard, the half-life of SARS-CoV-2 was longer than that of SARS-CoV-1. The longest viability of both viruses was on stainless steel and plastic; the estimated median half-life of SARS-CoV-2 was approximately 5.6 hours on stainless steel and 6.8 hours on plastic (Figure 1C). Estimated differences in the half-lives of the two viruses were small except for those on cardboard (Figure 1C). Individual replicate data were noticeably “noisier” (i.e., there was more variation in the experiment, resulting in a larger standard error) for cardboard than for other surfaces (Fig. S1 through S5), so we advise caution in interpreting this result.
We found that the stability of SARS-CoV-2 was similar to that of SARS-CoV-1 under the experimental circumstances tested. This indicates that differences in the epidemiologic characteristics of these viruses probably arise from other factors, including high viral loads in the upper respiratory tract and the potential for persons infected with SARS-CoV-2 to shed and transmit the virus while asymptomatic.3,4 Our results indicate that aerosol and fomite transmission of SARS-CoV-2 is plausible, since the virus can remain viable and infectious in aerosols for hours and on surfaces up to days (depending on the inoculum shed). These findings echo those with SARS-CoV-1, in which these forms of transmission were associated with nosocomial spread and super-spreading events,5 and they provide information for pandemic mitigation efforts.
A novel human coronavirus that is now named severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) (formerly called HCoV-19) emerged in Wuhan, China, in late 2019 and is now causing a pandemic.1 We analyzed the aerosol and surface stability of SARS-CoV-2 and compared it with SARS-CoV-1, the most closely related human coronavirus.2
We evaluated the stability of SARS-CoV-2 and SARS-CoV-1 in aerosols and on various surfaces and estimated their decay rates using a Bayesian regression model (see the Methods section in the Supplementary Appendix, available with the full text of this letter at NEJM.org). SARS-CoV-2 nCoV-WA1-2020 (MN985325.1) and SARS-CoV-1 Tor2 (AY274119.3) were the strains used. Aerosols (<5 μm) containing SARS-CoV-2 (105.25 50% tissue-culture infectious dose [TCID50] per milliliter) or SARS-CoV-1 (106.75-7.00 TCID50 per milliliter) were generated with the use of a three-jet Collison nebulizer and fed into a Goldberg drum to create an aerosolized environment. The inoculum resulted in cycle-threshold values between 20 and 22, similar to those observed in samples obtained from the upper and lower respiratory tract in humans.
Our data consisted of 10 experimental conditions involving two viruses (SARS-CoV-2 and SARS-CoV-1) in five environmental conditions (aerosols, plastic, stainless steel, copper, and cardboard). All experimental measurements are reported as means across three replicates.
SARS-CoV-2 remained viable in aerosols throughout the duration of our experiment (3 hours), with a reduction in infectious titer from 103.5 to 102.7 TCID50 per liter of air. This reduction was similar to that observed with SARS-CoV-1, from 104.3 to 103.5 TCID50 per milliliter (Figure 1A).
SARS-CoV-2 was more stable on plastic and stainless steel than on copper and cardboard, and viable virus was detected up to 72 hours after application to these surfaces (Figure 1A), although the virus titer was greatly reduced (from 103.7 to 100.6 TCID50 per milliliter of medium after 72 hours on plastic and from 103.7 to 100.6 TCID50 per milliliter after 48 hours on stainless steel). The stability kinetics of SARS-CoV-1 were similar (from 103.4 to 100.7 TCID50 per milliliter after 72 hours on plastic and from 103.6 to 100.6 TCID50 per milliliter after 48 hours on stainless steel). On copper, no viable SARS-CoV-2 was measured after 4 hours and no viable SARS-CoV-1 was measured after 8 hours. On cardboard, no viable SARS-CoV-2 was measured after 24 hours and no viable SARS-CoV-1 was measured after 8 hours (Figure 1A).
Both viruses had an exponential decay in virus titer across all experimental conditions, as indicated by a linear decrease in the log10TCID50 per liter of air or milliliter of medium over time (Figure 1B). The half-lives of SARS-CoV-2 and SARS-CoV-1 were similar in aerosols, with median estimates of approximately 1.1 to 1.2 hours and 95% credible intervals of 0.64 to 2.64 for SARS-CoV-2 and 0.78 to 2.43 for SARS-CoV-1 (Figure 1C, and Table S1 in the Supplementary Appendix). The half-lives of the two viruses were also similar on copper. On cardboard, the half-life of SARS-CoV-2 was longer than that of SARS-CoV-1. The longest viability of both viruses was on stainless steel and plastic; the estimated median half-life of SARS-CoV-2 was approximately 5.6 hours on stainless steel and 6.8 hours on plastic (Figure 1C). Estimated differences in the half-lives of the two viruses were small except for those on cardboard (Figure 1C). Individual replicate data were noticeably “noisier” (i.e., there was more variation in the experiment, resulting in a larger standard error) for cardboard than for other surfaces (Fig. S1 through S5), so we advise caution in interpreting this result.
We found that the stability of SARS-CoV-2 was similar to that of SARS-CoV-1 under the experimental circumstances tested. This indicates that differences in the epidemiologic characteristics of these viruses probably arise from other factors, including high viral loads in the upper respiratory tract and the potential for persons infected with SARS-CoV-2 to shed and transmit the virus while asymptomatic.3,4 Our results indicate that aerosol and fomite transmission of SARS-CoV-2 is plausible, since the virus can remain viable and infectious in aerosols for hours and on surfaces up to days (depending on the inoculum shed). These findings echo those with SARS-CoV-1, in which these forms of transmission were associated with nosocomial spread and super-spreading events,5 and they provide information for pandemic mitigation efforts.


Dit is niet best toch, kan je dus in je tuin het oplopen van iemand straten verderop etc. Nachtmerriequote:Op woensdag 18 maart 2020 10:43 schreef Momo het volgende:
Aerosol and Surface Stability of SARS-CoV-2 as Compared with SARS-CoV-1
A novel human coronavirus that is now named severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) (formerly called HCoV-19) emerged in Wuhan, China, in late 2019 and is now causing a pandemic.1 We analyzed the aerosol and surface stability of SARS-CoV-2 and compared it with SARS-CoV-1, the most closely related human coronavirus.2
We evaluated the stability of SARS-CoV-2 and SARS-CoV-1 in aerosols and on various surfaces and estimated their decay rates using a Bayesian regression model (see the Methods section in the Supplementary Appendix, available with the full text of this letter at NEJM.org). SARS-CoV-2 nCoV-WA1-2020 (MN985325.1) and SARS-CoV-1 Tor2 (AY274119.3) were the strains used. Aerosols (<5 μm) containing SARS-CoV-2 (105.25 50% tissue-culture infectious dose [TCID50] per milliliter) or SARS-CoV-1 (106.75-7.00 TCID50 per milliliter) were generated with the use of a three-jet Collison nebulizer and fed into a Goldberg drum to create an aerosolized environment. The inoculum resulted in cycle-threshold values between 20 and 22, similar to those observed in samples obtained from the upper and lower respiratory tract in humans.
Our data consisted of 10 experimental conditions involving two viruses (SARS-CoV-2 and SARS-CoV-1) in five environmental conditions (aerosols, plastic, stainless steel, copper, and cardboard). All experimental measurements are reported as means across three replicates.
SARS-CoV-2 remained viable in aerosols throughout the duration of our experiment (3 hours), with a reduction in infectious titer from 103.5 to 102.7 TCID50 per liter of air. This reduction was similar to that observed with SARS-CoV-1, from 104.3 to 103.5 TCID50 per milliliter (Figure 1A).
SARS-CoV-2 was more stable on plastic and stainless steel than on copper and cardboard, and viable virus was detected up to 72 hours after application to these surfaces (Figure 1A), although the virus titer was greatly reduced (from 103.7 to 100.6 TCID50 per milliliter of medium after 72 hours on plastic and from 103.7 to 100.6 TCID50 per milliliter after 48 hours on stainless steel). The stability kinetics of SARS-CoV-1 were similar (from 103.4 to 100.7 TCID50 per milliliter after 72 hours on plastic and from 103.6 to 100.6 TCID50 per milliliter after 48 hours on stainless steel). On copper, no viable SARS-CoV-2 was measured after 4 hours and no viable SARS-CoV-1 was measured after 8 hours. On cardboard, no viable SARS-CoV-2 was measured after 24 hours and no viable SARS-CoV-1 was measured after 8 hours (Figure 1A).
Both viruses had an exponential decay in virus titer across all experimental conditions, as indicated by a linear decrease in the log10TCID50 per liter of air or milliliter of medium over time (Figure 1B). The half-lives of SARS-CoV-2 and SARS-CoV-1 were similar in aerosols, with median estimates of approximately 1.1 to 1.2 hours and 95% credible intervals of 0.64 to 2.64 for SARS-CoV-2 and 0.78 to 2.43 for SARS-CoV-1 (Figure 1C, and Table S1 in the Supplementary Appendix). The half-lives of the two viruses were also similar on copper. On cardboard, the half-life of SARS-CoV-2 was longer than that of SARS-CoV-1. The longest viability of both viruses was on stainless steel and plastic; the estimated median half-life of SARS-CoV-2 was approximately 5.6 hours on stainless steel and 6.8 hours on plastic (Figure 1C). Estimated differences in the half-lives of the two viruses were small except for those on cardboard (Figure 1C). Individual replicate data were noticeably “noisier” (i.e., there was more variation in the experiment, resulting in a larger standard error) for cardboard than for other surfaces (Fig. S1 through S5), so we advise caution in interpreting this result.
We found that the stability of SARS-CoV-2 was similar to that of SARS-CoV-1 under the experimental circumstances tested. This indicates that differences in the epidemiologic characteristics of these viruses probably arise from other factors, including high viral loads in the upper respiratory tract and the potential for persons infected with SARS-CoV-2 to shed and transmit the virus while asymptomatic.3,4 Our results indicate that aerosol and fomite transmission of SARS-CoV-2 is plausible, since the virus can remain viable and infectious in aerosols for hours and on surfaces up to days (depending on the inoculum shed). These findings echo those with SARS-CoV-1, in which these forms of transmission were associated with nosocomial spread and super-spreading events,5 and they provide information for pandemic mitigation efforts.
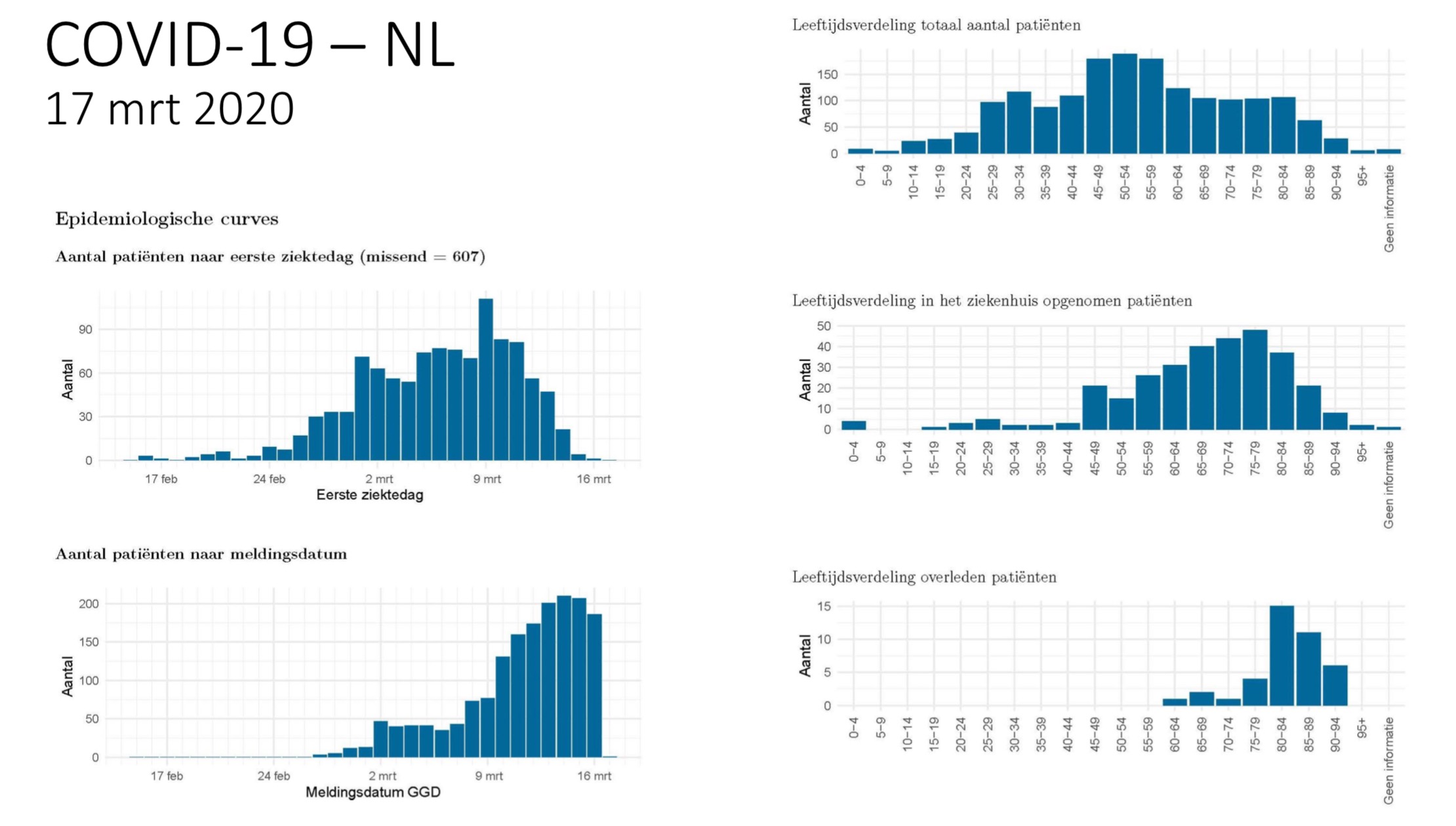
[ afbeelding ]


Mutations can reveal how the coronavirus moves—but they’re easy to overinterpret
mmediately after Christian Drosten published a genetic sequence of the novel coronavirus online on 28 February, he took to Twitter to issue a warning. As the virus has raced around the world, more than 350 genome sequences have been shared on the online platform GISAID. They hold clues to how the new virus, named severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), is spreading and evolving. But because the sequences represent a tiny fraction of cases and show few telltale differences, they are easy to overinterpret, as Drosten realized.
A virologist at the Charitť University Hospital in Berlin, he had sequenced the virus from a German patient infected with COVID-19 in Italy. The genome looked similar to that of a virus found in a patient in Munich, the capital of Bavaria, more than 1 month earlier; both shared three mutations not seen in early sequences from China. Drosten realized this could give rise to the idea that the Italian outbreak was “seeded” by the one in Bavaria, which state public health officials said had been quashed by tracing and quarantining all contacts of the 14 confirmed cases. But he thought it was just as likely that a Chinese variant carrying the three mutations had taken independent routes to both countries. The newly sequenced genome “is not sufficient to claim a link between Munich and Italy,” Drosten tweeted.
His warning went unheeded. A few days later, Trevor Bedford of the Fred Hutchinson Cancer Research Center, who analyzes the stream of viral genomes and discusses them in Twitter threads, wrote that the pattern suggested the outbreak in Bavaria had not been contained after all, and appeared to have led to the Italian outbreak. The analysis spread widely. Technology Review asserted that “the Munich event could be linked to a decent part of the overall European outbreak” and Twitter users called on Germany to apologize. (This Science correspondent retweeted Bedford’s thread as well.)
irologist Eeva Broberg of the European Centre for Disease Prevention and Control agrees with Drosten that there are more plausible scenarios for how the disease reached northern Italy than an undetected spread from Bavaria. Other scientists say Bedford jumped the gun as well. “I have to kick his butt a bit for this,” says Richard Neher, a computational biologist at the University of Basel who works with Bedford. “It’s a cautionary tale,” says Andrew Rambaut, a molecular evolutionary biologist at the University of Edinburgh. “There is no way you can make that claim just from the phylogeny alone.” Bedford later clarified he believed it was equally plausible there had been two separate introductions from China. “I think I should have been more careful with that Twitter thread,” he says.
It was a case study in the power and pitfalls of real-time analysis of viral genomes. “This is an incredibly important disease. We need to understand how it is moving,” says Bette Korber, a biologist at Los Alamos National Laboratory who is also studying the genome of SARS-CoV-2. “With very limited evolution during the outbreak, [these researchers] are doing what they can and they are making suggestions, which I think at this point should be taken as suggestions.”
The sequence data were most informative early on, says Kristian Andersen, a computational biologist at Scripps Research. The very first sequence, in early January, answered the most basic question: What pathogen is causing the disease? The ones that followed were almost identical, strongly suggesting there was a single introduction from an animal into the human population. If the virus had jumped the species barrier multiple times, scientists would see more variety among the first human cases.
Now, more diversity is emerging. Like all viruses, SARS-CoV-2 evolves over time through random mutations, only some of which are caught and corrected by the virus’s error correction machinery. Over the length of its 30,000-base-pair genome, SARS-CoV-2 accumulates an average of about one to two mutations per month, Rambaut says. “It’s about two to four times slower than the flu,” he says. Using these little changes, researchers can draw up phylogenetic trees, much like family trees. They can also make connections between different cases of COVID-19 and gauge whether there might be undetected spread of the virus.
For instance, when researchers sequenced the second virus genome in Washington—from a teenager diagnosed with COVID-19 on 27 February—it looked like a direct descendant of the first genome, a case found 6 weeks earlier, that had acquired three further mutations. Bedford tweeted that he considered it “highly unlikely” that the two genomes came from separate introductions. “I believe we are facing an already substantial outbreak in Washington State that was not detected until now,” he wrote. That analysis turned out to be correct: Washington has now reported more than 100 cases and 15 deaths and additional genomes from other patients have bolstered the link. In this case, Bedford’s hypothesis was much stronger because the two patients both came from Snohomish County, Rambaut says: “It’s very unlikely that this highly related virus would travel to exactly the same town in Washington,” he says.
Few other firm conclusions about the virus’s spread have emerged, in part because the wealth of genomes is still a tiny sample of the more than 100,000 cases worldwide. Although China accounts for 80% of all COVID-19 cases, only one-third of the published genomes are from China—and very few of them are from later cases. And because it’s early in the outbreak, most genomes are still very similar, which makes it hard to draw conclusions. “We just have this handful of mutations, which makes these groupings so ambiguous,” Neher says. “As the outbreak unfolds, we expect to see more and more diversity and more clearly distinct lineages,” he says. “And then it will become easier and easier to actually put things together.”
Scientists will also be scouring the genomic diversity for mutations that might change how dangerous the pathogen is or how fast it spreads. There, too, caution is warranted. A paper published by Lu Jian of Peking University and colleagues on 3 March in the journal National Science Review analyzed 103 virus genomes and argued that they fell into one of two distinct types, named S and L, distinguished by two mutations. Because 70% of sequenced SARS-CoV-2 genomes belong to L, the newer type, the authors concluded that virus has evolved to become more aggressive and to spread faster.
But they lack evidence, Rambaut says. “What they’ve done is basically seen these two branches and said, ‘That one is bigger, [so that virus] must be more virulent or more transmissible,’” he says. However, just because a virus is exported and leads to a large outbreak elsewhere does not mean it is behaving differently: “One of these lineages is going to be bigger than the other just by chance.” Some researchers have called for the paper to be retracted. “The claims made in it are clearly unfounded and risk spreading dangerous misinformation at a crucial time in the outbreak,” four scientists at the University of Glasgow wrote in a response published on www.virological.org. (In a response, Lu wrote the four had misunderstood his study.)
Most genomic changes don’t alter the virus’s behavior, Drosten says. The only way to confirm that a mutation has an effect is to study it in cell cultures or animal models and show, for instance, that it has become better at entering cells or transmitting, he says. And if the virus does change in an important way, it could go either way, making it more or less dangerous. In 2018, Drosten’s group published a paper showing that early in the SARS outbreak of 2002–03, that virus lost a small chunk of its genome, 29 base pairs in one gene. Adding those base pairs back in the lab made the virus much better at replicating in several cell culture models.
It might seem strange that a mutation that weakens the virus would become established, but that can happen when it has just entered the human population and isn’t competing with strains lacking the mutation, Drosten says. “Sadly, this new virus doesn’t have that deletion,” he adds
mmediately after Christian Drosten published a genetic sequence of the novel coronavirus online on 28 February, he took to Twitter to issue a warning. As the virus has raced around the world, more than 350 genome sequences have been shared on the online platform GISAID. They hold clues to how the new virus, named severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), is spreading and evolving. But because the sequences represent a tiny fraction of cases and show few telltale differences, they are easy to overinterpret, as Drosten realized.
A virologist at the Charitť University Hospital in Berlin, he had sequenced the virus from a German patient infected with COVID-19 in Italy. The genome looked similar to that of a virus found in a patient in Munich, the capital of Bavaria, more than 1 month earlier; both shared three mutations not seen in early sequences from China. Drosten realized this could give rise to the idea that the Italian outbreak was “seeded” by the one in Bavaria, which state public health officials said had been quashed by tracing and quarantining all contacts of the 14 confirmed cases. But he thought it was just as likely that a Chinese variant carrying the three mutations had taken independent routes to both countries. The newly sequenced genome “is not sufficient to claim a link between Munich and Italy,” Drosten tweeted.
His warning went unheeded. A few days later, Trevor Bedford of the Fred Hutchinson Cancer Research Center, who analyzes the stream of viral genomes and discusses them in Twitter threads, wrote that the pattern suggested the outbreak in Bavaria had not been contained after all, and appeared to have led to the Italian outbreak. The analysis spread widely. Technology Review asserted that “the Munich event could be linked to a decent part of the overall European outbreak” and Twitter users called on Germany to apologize. (This Science correspondent retweeted Bedford’s thread as well.)
irologist Eeva Broberg of the European Centre for Disease Prevention and Control agrees with Drosten that there are more plausible scenarios for how the disease reached northern Italy than an undetected spread from Bavaria. Other scientists say Bedford jumped the gun as well. “I have to kick his butt a bit for this,” says Richard Neher, a computational biologist at the University of Basel who works with Bedford. “It’s a cautionary tale,” says Andrew Rambaut, a molecular evolutionary biologist at the University of Edinburgh. “There is no way you can make that claim just from the phylogeny alone.” Bedford later clarified he believed it was equally plausible there had been two separate introductions from China. “I think I should have been more careful with that Twitter thread,” he says.
It was a case study in the power and pitfalls of real-time analysis of viral genomes. “This is an incredibly important disease. We need to understand how it is moving,” says Bette Korber, a biologist at Los Alamos National Laboratory who is also studying the genome of SARS-CoV-2. “With very limited evolution during the outbreak, [these researchers] are doing what they can and they are making suggestions, which I think at this point should be taken as suggestions.”
The sequence data were most informative early on, says Kristian Andersen, a computational biologist at Scripps Research. The very first sequence, in early January, answered the most basic question: What pathogen is causing the disease? The ones that followed were almost identical, strongly suggesting there was a single introduction from an animal into the human population. If the virus had jumped the species barrier multiple times, scientists would see more variety among the first human cases.
Now, more diversity is emerging. Like all viruses, SARS-CoV-2 evolves over time through random mutations, only some of which are caught and corrected by the virus’s error correction machinery. Over the length of its 30,000-base-pair genome, SARS-CoV-2 accumulates an average of about one to two mutations per month, Rambaut says. “It’s about two to four times slower than the flu,” he says. Using these little changes, researchers can draw up phylogenetic trees, much like family trees. They can also make connections between different cases of COVID-19 and gauge whether there might be undetected spread of the virus.
For instance, when researchers sequenced the second virus genome in Washington—from a teenager diagnosed with COVID-19 on 27 February—it looked like a direct descendant of the first genome, a case found 6 weeks earlier, that had acquired three further mutations. Bedford tweeted that he considered it “highly unlikely” that the two genomes came from separate introductions. “I believe we are facing an already substantial outbreak in Washington State that was not detected until now,” he wrote. That analysis turned out to be correct: Washington has now reported more than 100 cases and 15 deaths and additional genomes from other patients have bolstered the link. In this case, Bedford’s hypothesis was much stronger because the two patients both came from Snohomish County, Rambaut says: “It’s very unlikely that this highly related virus would travel to exactly the same town in Washington,” he says.
Few other firm conclusions about the virus’s spread have emerged, in part because the wealth of genomes is still a tiny sample of the more than 100,000 cases worldwide. Although China accounts for 80% of all COVID-19 cases, only one-third of the published genomes are from China—and very few of them are from later cases. And because it’s early in the outbreak, most genomes are still very similar, which makes it hard to draw conclusions. “We just have this handful of mutations, which makes these groupings so ambiguous,” Neher says. “As the outbreak unfolds, we expect to see more and more diversity and more clearly distinct lineages,” he says. “And then it will become easier and easier to actually put things together.”
Scientists will also be scouring the genomic diversity for mutations that might change how dangerous the pathogen is or how fast it spreads. There, too, caution is warranted. A paper published by Lu Jian of Peking University and colleagues on 3 March in the journal National Science Review analyzed 103 virus genomes and argued that they fell into one of two distinct types, named S and L, distinguished by two mutations. Because 70% of sequenced SARS-CoV-2 genomes belong to L, the newer type, the authors concluded that virus has evolved to become more aggressive and to spread faster.
But they lack evidence, Rambaut says. “What they’ve done is basically seen these two branches and said, ‘That one is bigger, [so that virus] must be more virulent or more transmissible,’” he says. However, just because a virus is exported and leads to a large outbreak elsewhere does not mean it is behaving differently: “One of these lineages is going to be bigger than the other just by chance.” Some researchers have called for the paper to be retracted. “The claims made in it are clearly unfounded and risk spreading dangerous misinformation at a crucial time in the outbreak,” four scientists at the University of Glasgow wrote in a response published on www.virological.org. (In a response, Lu wrote the four had misunderstood his study.)
Most genomic changes don’t alter the virus’s behavior, Drosten says. The only way to confirm that a mutation has an effect is to study it in cell cultures or animal models and show, for instance, that it has become better at entering cells or transmitting, he says. And if the virus does change in an important way, it could go either way, making it more or less dangerous. In 2018, Drosten’s group published a paper showing that early in the SARS outbreak of 2002–03, that virus lost a small chunk of its genome, 29 base pairs in one gene. Adding those base pairs back in the lab made the virus much better at replicating in several cell culture models.
It might seem strange that a mutation that weakens the virus would become established, but that can happen when it has just entered the human population and isn’t competing with strains lacking the mutation, Drosten says. “Sadly, this new virus doesn’t have that deletion,” he adds


COVID-19 coronavirus epidemic has a natural origin
The novel SARS-CoV-2 coronavirus that emerged in the city of Wuhan, China, last year and has since caused a large scale COVID-19 epidemic and spread to more than 70 other countries is the product of natural evolution, according to findings published today in the journal Nature Medicine.
Kristian G. Andersen, Andrew Rambaut, W. Ian Lipkin, Edward C. Holmes, Robert F. Garry. The proximal origin of SARS-CoV-2. Nature Medicine, 2020; DOI: 10.1038/s41591-020-0820-9
Median incubation period for COVID-19
A new study calculates that the median incubation period for COVID-19 is just over 5 days and that 97.5% of people who develop symptoms will do so within 11.5 days of infection.
Stephen A. Lauer, Kyra H. Grantz, Qifang Bi, Forrest K. Jones, Qulu Zheng, Hannah R. Meredith, Andrew S. Azman, Nicholas G. Reich, Justin Lessler. The Incubation Period of Coronavirus Disease 2019 (COVID-19) From Publicly Reported Confirmed Cases: Estimation and Application. Annals of Internal Medicine, 2020; DOI: 10.7326/M20-0504
New coronavirus stable for hours on surfaces
Ah die stond al hierboven zie ik.
The novel SARS-CoV-2 coronavirus that emerged in the city of Wuhan, China, last year and has since caused a large scale COVID-19 epidemic and spread to more than 70 other countries is the product of natural evolution, according to findings published today in the journal Nature Medicine.
Kristian G. Andersen, Andrew Rambaut, W. Ian Lipkin, Edward C. Holmes, Robert F. Garry. The proximal origin of SARS-CoV-2. Nature Medicine, 2020; DOI: 10.1038/s41591-020-0820-9
Median incubation period for COVID-19
A new study calculates that the median incubation period for COVID-19 is just over 5 days and that 97.5% of people who develop symptoms will do so within 11.5 days of infection.
Stephen A. Lauer, Kyra H. Grantz, Qifang Bi, Forrest K. Jones, Qulu Zheng, Hannah R. Meredith, Andrew S. Azman, Nicholas G. Reich, Justin Lessler. The Incubation Period of Coronavirus Disease 2019 (COVID-19) From Publicly Reported Confirmed Cases: Estimation and Application. Annals of Internal Medicine, 2020; DOI: 10.7326/M20-0504
New coronavirus stable for hours on surfaces
Ah die stond al hierboven zie ik.
|
|
| Forum Opties | |
|---|---|
| Forumhop: | |
| Hop naar: | |